Part:BBa_K131000:Design
ColE2 Operon
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 2228
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Design Notes
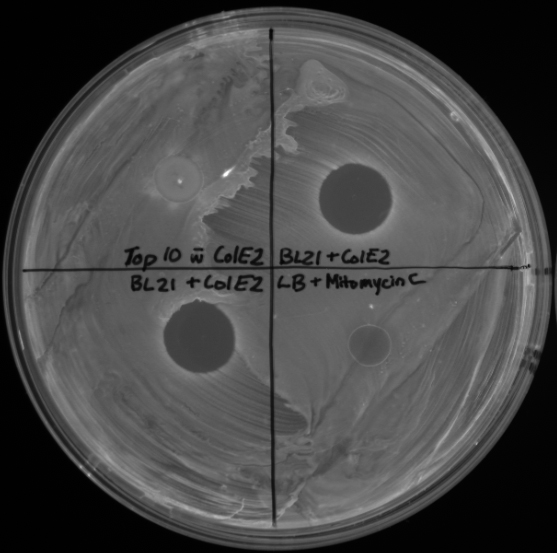
Since the ColE2 operon is under the control of the SOS promoter, a RecA positive strain of bacteria must be used for expression. For this purpose we used [http://www.emdbiosciences.com/product/69449 BL21] cells from Novagen, which are a RecA positive E. coli strain. We took BL21 strains containing the ColE2 operon and grew cultures (5 mL) overnight. Since we want to induce an SOS response, we used the antibiotic [http://www.sigmaaldrich.com/catalog/search/ProductDetail/SIGMA/M4287 Mitomycin C] from Sigma. For a 5 mL culture, this would require the addition of 10uL of 0.5mg/mL mitomycin C. This was to achieve Sigma's recommendation to achieve a final mitomycin C concentration of 1ug/mL. Inducing the cultures 2 hours before spotting onto a fresh lawn of bacteria was adequate and demonstrated a comparable response to cultures induced 4 hours before.
It was important that the lawn of bacteria is freshly prepared before you spot on the induced BL21 cells. There was no noticeable difference between supernatant and resuspended cells in terms of clearing.
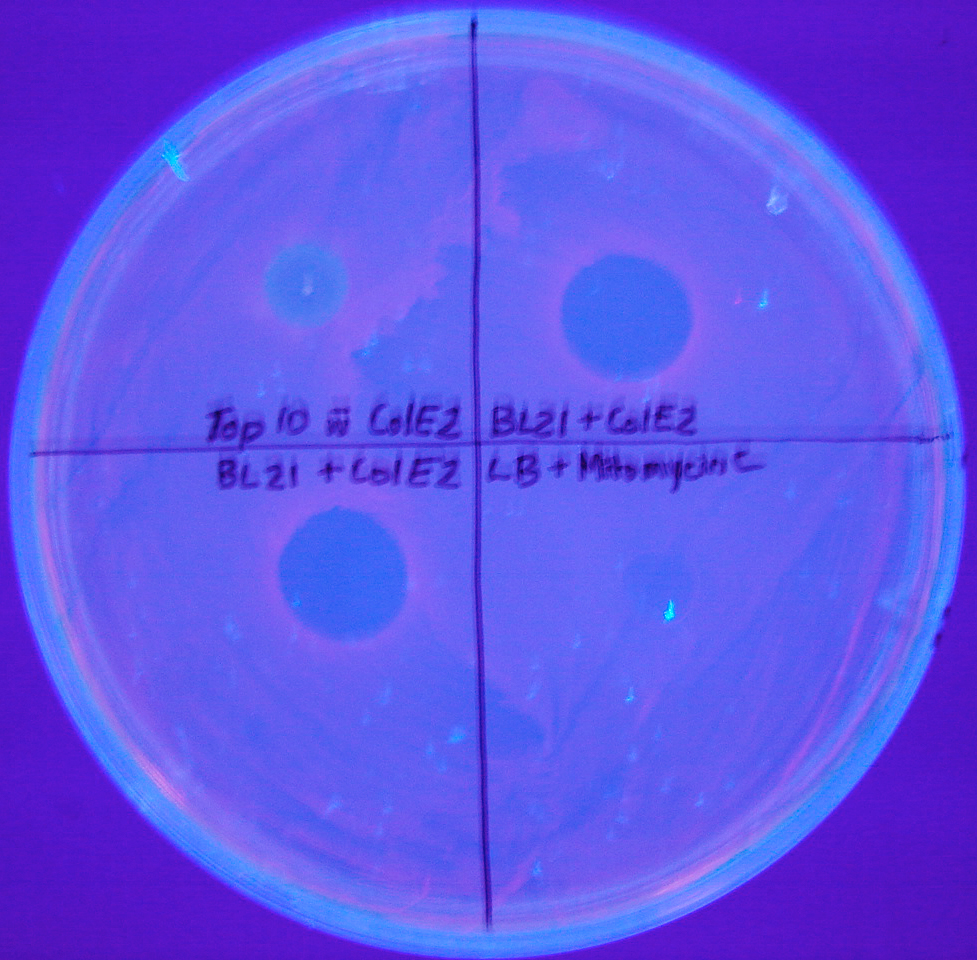
In the pictures below where induced BL21 cultures were spotted onto a lawn of RFP bacteria, note the controls. In Top10 E. coli (upper left quadrant), there is no clearing. Both the upper right and bottom left quadrants were BL21 strains induced with mitomycin C 2 hours before spotting. The bottom right quadrant is LB and mitomycin C, showing that the antibiotic itself was not creating the clearing.
The third picture in the upper left quadrant has as a control BL21 containing the plasmid with ColE2, but not induced. Also, the induced cultures on this plate were spotted 2 hours and 4 hours after induction.
Pictures
Source
This part was cloned from a plasmid containing the operon which was received from UC Berkeley.

 1 Registry Star
1 Registry Star